Yushan Xia (HKU)
- Department of Molecular Genetics, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Groningen, Netherlands
- State Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Molecular Microbiology and Technology of the Ministry of Education, Department of Microbiology, College of Life Sciences, Nankai University, Tianjin, P.R. China.
- Department of Chemistry, The University of Hong Kong, Hong Kong, P.R. China.
(E-mail: yushan93@hku.hk)
Abstract.
The emergence and rapid spread of multi-drug resistant (MDR) bacteria pose a serious threat to global healthcare systems. There is an urgent need for new antibacterial substances and/or new treatment strategies to deal with the infections by MDR pathogens, especially the Gram-negative pathogens, which are the most challenging group. In this study, we investigate a “Trojan horse” strategy that employs the anti-Helicobacter pylori drug bismuth subsalicylate and related bismuth compound salts. We found that these compounds specifically disrupt Pseudomonas aeruginosa iron metabolism, inhibit the activity of the iron-sulfur cluster-dependent respiration complex, dissipate the proton motive force, reduce ATP production and inhibit the activity of efflux pumps. As a consequence of these alterations and despite the fact that bismuth alone is completely inactive, the efficacy of tetracycline-related antibiotics against P. aeruginosa in vitro is restored, and the development of resistance to tetracycline-related antibiotics is also impaired. In addition, we found that the combination of bismuth compounds and tetracyclines, used at non-toxic concentrations for human cells, enhanced the elimination of established biofilms. Finally, we demonstrate the activity of this combination in vitro against 124 clinical P. aeruginosa isolates. Our study provides a realistic therapeutical option for combining the FDA approved bismuth-based compounds with tetracycline-related antibiotics to combat infections caused by P. aeruginosa.
Simranjeet Kaur (HKBU)
1 Department of Chemistry, Hong Kong Baptist University, Kowloon Tong, Hong Kong SAR, China
2 The Department of Chemistry and the Ineos Oxford Institute for Antimicrobial Research, Chemistry Research Laboratory, University of Oxford, 12 Mansfield Road, Oxford, OX1 3TA, United Kingdom.
(E-mail: 20481187@life.hkbu.edu.hk)
Abstract.
AlkB homologue 5 (ALKBH5) belongs to the family of Fe(II) and 2-oxoglutarare (2OG) dependent oxygenases and catalyses the oxidative demethylation of N6-methyladenosine (m6A),1 a post-transcriptional RNA modification with critical roles in gene regulation and disease progression.2 It is one of the only two discovered human m6A RNA oxidizing enzymes, along with the fat mass and obesity associated protein (FTO). ALKBH5 has gained wide attention as a potential target for cancer treatment; ALKBH5 mediated m6A demethylation is linked with different types of cancers including glioblastoma.3 However, the underlying molecular mechanisms of substrate recognition and demethylation by ALKBH5 are yet to be understood due to the lack of a structure of ALKBH5 in complex with its RNA subtrate. Here, we present three crystal structures of ALKBH5 in complex with an m6A-ssRNA 8-mer substrate and supporting biochemical studies. Our studies demonstrate how, unlike FTO which produces N6-hydroxymethyladenosine (hm6A) along with demethylated adenosine, ALKBH5 effectively produces a demethylated adenosine by catalyzing the fragmentation of its proposed nascent hemiaminal (hm6A) intermediate.4 We discovered that the 5′-3′ direction of the single-stranded RNA substrate when bound to the active site of ALKBH5 is opposite to substrates bound to other AlkB subfamily members, including single-stranded DNA bound to FTO4. Our structural and biochemical findings also provided insights into the substrate preferences of ALKBH5 with a consensus (A/G)m6AC sequence motif. Based on our structural and biochemical data, we proposed a catalytic mechanism involving a proton shuttle network for the demethylation of m6A by ALKBH54.
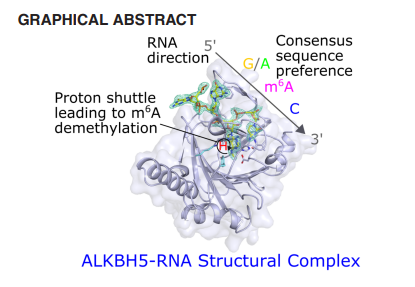
(Adapted from Kaur, S.; Tam, N. Y.; McDonough, M. A.; Schofield, C. J.; Aik, W. S., Nucleic Acids Research 2022, 50 (7), 4148-4160.)
References
(1) Zheng, G.; Dahl, J. A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C. J.; Vågbø, C. B.; Shi, Y.; Wang, W.-L.; Song, S.-H., Molecular cell 2013, 49 (1), 18-29.
(2) Yang, Y.; Hsu, P. J.; Chen, Y.-S.; Yang, Y.-G., Cell research 2018, 28 (6), 616-624.
(3) Zhang, S.; Zhao, B. S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E. P.; Xie, K.; Bögler, Cancer cell 2017, 31 (4), 591-606. e596.
(4) Kaur, S.; Tam, N. Y.; McDonough, M. A.; Schofield, C. J.; Aik, W. S., Nucleic Acids Research 2022, 50 (7), 4148-4160.
Winnie Tam (HKU)
The University of Hong Kong
(E-mail: twl3523@connect.hku.hk)
Abstract.
Iron-sulfur (Fe-S) clusters are essential in DNA replication and genome integrity maintenance[1-3]. The coordination of Fe-S clusters is crucial for DNA replicative and repair enzymes to carry out their physiological functions. As 4Fe-4S clusters are involved actively in DNA replication and repair, mutations in Fe-S protein may lead to genome instability and cancer formation[4, 5]. While 4Fe-4S protein is susceptible to degradation in oxic environment, we have developed a combinatorial approach to identify novel cancer-associated 4Fe-4S protein in silico. We have analyzed information from various data bases, cysteine geometry from RSCB PDB, signature motif of 4Fe-4S assembly from UniProt and cancer-associated cysteine mutant from COSMIC. Out of 73101 human proteome in UniProt, 22 proteins, including know cancer-associated 4Fe-4S protein MUTYH, fulfil all three criteria. Our results suggest intriguing possibility of 4Fe-4S binding site in histone lysine methyltransferase as well as other receptors and growth factors. This provides insight for further in vitro studies design to prove the existence of 4Fe-4S cluster. Better understanding on these cancer-associated protein would be beneficial to cancer prognosis and treatment.
References
- Paul, V.D. and R. Lill, Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim Biophys Acta, 2015. 1853(6): p. 1528-39.
- Weiner, B.E., et al., An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J Biol Chem, 2007. 282(46): p. 33444-51.
- White, M.F. and M.S. Dillingham, Iron-sulphur clusters in nucleic acid processing enzymes. Curr Opin Struct Biol, 2012. 22(1): p. 94-100.
- Paulo, P., et al., Targeted next generation sequencing identifies functionally deleterious germline mutations in novel genes in early-onset/familial prostate cancer. PLoS Genet, 2018. 14(4): p. e1007355.
- McDonnell, K.J., et al., A human MUTYH variant linking colonic polyposis to redox degradation of the [4Fe4S](2+) cluster. Nat Chem, 2018. 10(8): p. 873-880.
